Our Science
Many high-value undrugged targets lack a ligandable conformation due to a high degree of disorder, leaving them intractable to small-molecule drug discovery. In order to address the substantial unmet clinical need that persists in many therapeutic areas such as cancer, neurodegenerative and metabolic disease, it is essential to systematically expand the druggable proteome.
Tremendous potential lies in the protein interactome, constituted of an estimated 130,000 to 650,000 protein complexes. Successes in targeting individual protein-protein interactions reported so far have required great efforts to develop effective compounds in each individual case. Ambagon holds the key to a modular strategy, enabling therapeutic intervention in a broad spectrum of disease pathways.
Through our team’s deep scientific expertise in the structure and interactions of 14-3-3 adaptor proteins, we developed a platform to deliver intrinsically disordered proteins to drug development. With an estimated 3,000 clients across the interactome, many of which are difficult to drug but with critical roles in disease, 14-3-3 allows for leveraging its endogenous regulatory function by small molecules.
interactome with more
than 3,000 interactions
14-3-3 Biology
14-3-3 proteins are adaptor proteins that play a key role in a large number of disease-related pathways. The seven 14-3-3 proteins form complexes with specific phosphorylated sequences in disordered domains of their partner proteins, affecting the functions of these clients to regulate cellular processes such as gene expression, signal transduction, protein trafficking and apoptosis. Many disease-relevant proteins are 14-3-3 clients, such as the cancer-driving Raf kinases, FOXO transcription factors, p53 tumor suppressor, and neurological disease targets such as tau, alpha synuclein, and the LRRK2 kinase.
How 14-3-3 Works
14-3-3 proteins interact with their clients in a specific binding groove through recognition of phosphorylated serine- or threonine-containing motifs, conferring order upon disordered domains and creating the structure necessary for small molecule recognition. 14-3-3:client binding is sufficiently variable that each one creates structurally unique pockets within the binding groove that can be occupied by selective small-molecule stabilizers/molecular glues. By stabilizing the naturally occurring 14-3-3:client interaction, these small-molecule drugs can enhance desired outcomes of client binding, such as sequestration away from the site of disease-driving activity to inhibit function, trafficking to a desired site of activity to restore function, enhancing or stabilizing activity, or promoting degradation of disease-driving clients.
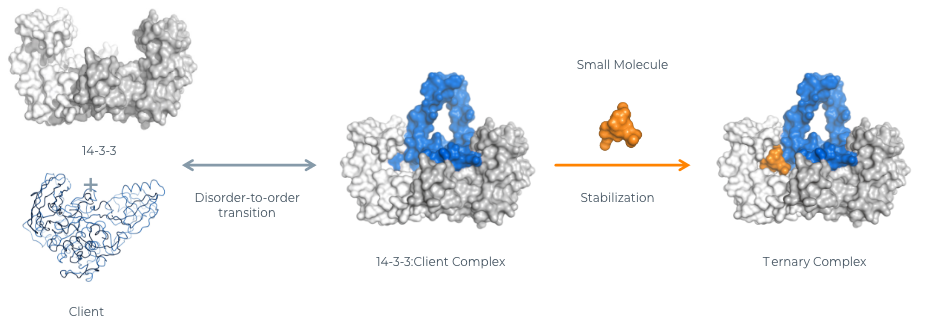
14-3-3 Proof of Concept and Possibilities
Natural compounds, such as the fungal metabolite Fusicoccin-A, are known to stabilize 14-3-3:client interactions and have inspired the creation of synthetic stabilizers that are able to constitute the same effect. These efforts resulted in a systematic understanding of the 14-3-3:client interface. Small-molecule drugs based on various chemotypes — from complex natural products to molecular fragments — can now be deployed to stabilize a range of these complexes. Our team has leveraged its deep understanding of 14-3-3 structure and protein-protein interactions to create a modular platform for discovering these compounds.
Selected Publications
14-3-3 and Carbohydrate Response Element Binding Protein (ChREBP)
Nature Communications, August 7, 2020
Site-directed fragment-based screening for the discovery of protein–protein interaction stabilizers
14-3-3 and Estrogen Receptor alpha (ERα)
American Chemical Society, February 1, 2019
Fragment-based differential targeting of PPI stabilizer interfaces
14-3-3 and p53 or TAZ
American Chemical Society, June 5, 2020
14-3-3, ERα and Fusicoccin
PNAS, May 15, 2013
American Chemical Society, March 16, 2023
Nature Communications, June 23, 2022